bleties
IES retention analysis from PacBio CCS reads
Blepharisma Toolbox for Interspersed DNA Elimination Studies (BleTIES)

BleTIES is a tool for prediction and targeted assembly of internally eliminated sequences (IESs) in ciliate genomes, using single-molecule long read sequencing. The design and name of the software was inspired by ParTIES.
Input data
- Ciliate MAC genome assembly, Fasta format.
- Long read library (PacBio subreads or CCS, Nanopore reads) mapping onto that assembly, sorted/indexed BAM format; mapper should report valid CIGAR string and NM tag. The mapping is assumed to be accurate.
Installation
Install released version with Conda
The released versions are distributed via Bioconda, and can be installed with Conda:
# Create new environment called "bleties"
conda create -c conda-forge -c bioconda -n bleties bleties
# Activate environment
conda activate bleties
# Check version and view help message
bleties --version
bleties --help
# Run tests
python -m unittest -v bleties.TestModule
Install development version
If you want to test the latest development version, clone this Git repository, then install with pip.
Dependencies are specified as a Conda environment YAML file env.yaml. Create a
Conda environment with the specified dependencies, then install with pip:
git clone git@github.com:Swart-lab/bleties.git
cd bleties
conda env create -f env.yaml -n bleties_dev
conda activate bleties_dev
pip install .
Run tests after installation:
python -m unittest -v bleties.TestModule
Usage
To list input arguments and options and their respective usage, use the -h or
--help option, with or without the subworkflow names:
bleties --help
bleties milraa --help
bleties miser --help
bleties milret --help
bleties milcor --help
bleties miltel --help
bleties insert --help
In addition there are two scripts for plotting the output from the MILRAA and
MILCOR modules. See the --help messages for usage instructions:
milcor_plot.py --help
milraa_plot.py --help
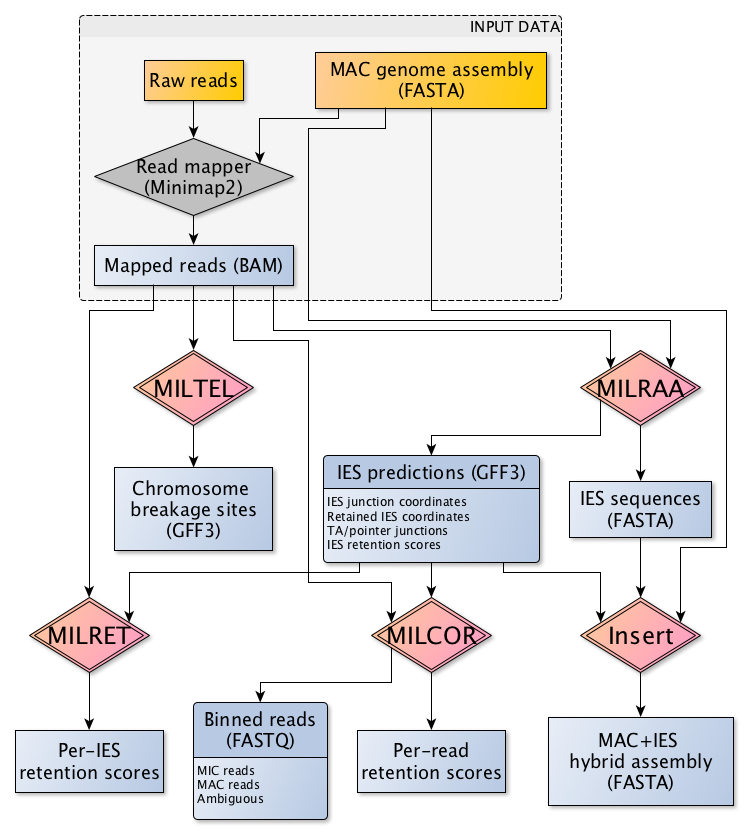
Outline of workflow
Refer to the individual module pages for further information.
- MILRAA – Identify putative IESs and IES junctions from a mapping of long reads (uncorrected PacBio or Nanopore, or PacBio CCS reads) to reference genome assembly.
- MISER – Screen for potentially erroneous IES calls, to curate the list of putative IES junctions (experimental!).
- MILRET – Use curated IES junctions, or IES junction coordinates from third-party tools, to calculate IES retention scores from mapping of CCS reads to a reference MAC (IES-) assembly.
- MILCOR – Calculate per-read IES retention scores and bin reads as MIC/MAC in origin, from mapping of CCS reads to MAC assembly.
- MILTEL – Identify potential chromosome breakage sites with telomere addition, from mapping of CCS reads to MAC assembly.
- Insert – Utility for inserting IES sequences from a MAC reference genome to generate MAC+IES hybrid sequence. Also can perform the reverse operation.
Citations
BleTIES is research software. Please cite us if you use the software in a publication.
Brandon K. B. Seah, Estienne C. Swart. (2021) BleTIES: Annotation of natural genome editing in ciliates using long read sequencing. Bioinformatics btab613; doi: https://doi.org/10.1093/bioinformatics/btab613

